Parallelizing cGAs
In the following points we will discuss some parallel cGAs
designed for working in massively parallel computers. In these algorithms, each
processing element processes a single individual:
- ASPARAGOS was introduced by Gorges-Schleuter
[Gorg89a,
Gorg89b]
and Mühlenbein [Mühl89a,
Mühl89b].
ASPARAGOS used a population
structure that resembles a ladder with the upper and lower ends tied
together. ASPARAGOS was used to solve some difficult
combinatorial optimization problems with great success. Later,
a linear population structure was used [Gorg91],
and different mating strategies were compared [Gorg92].
ASPARAGOS uses a local hillclimbing
algorithm to improve the fitness of the individuals in its population.
This makes it difficult to isolate the contribution of the spatial structure
of the population to the search quality. However, all the empirical results
show that ASPARAGOS is a very effective optimization
tool.
- Manderick and Spiessens [MS89]
implemented a fine-grained parallel GA with the population
distributed on a 2-D grid. Selection and mating were only possible
within a neighborhood, and the authors observed that the performance of the
algorithm degraded as the size of the neighborhood increased. On the extreme,
if the size of the neighborhood was big enough, this parallel GA was equivalent
to a single panmistic population. The authors analyzed this algorithm theoretically
in subsequent years [SM90,
SM91], and
their analysis showed that for a deme size s and
a string length l, the time complexity of the algorithm
was
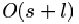 or
or
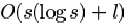 time
steps, depending on the selection scheme used.
time
steps, depending on the selection scheme used.
- Schwehm [Schw92]
[Schw94] compared
the effect of using different structures on the population of fine-grained
GAs. In these papers, a graph partitioning problem was used as a benchmark
to test different population structures simulated on a MasPar MP-1 computer.
Among the structures tested there were a ring, a torus,
a 16 x 8 x 8 cube a 4 x 4 x 4 x 4 x 4 hypercube, and a 10-D binary hypercube.
The algorithm with the torus structure converged faster than the other algorithms.
The implementation of the algorithm is discussed in [Schw93].
- Thompson et al. developed in [TBK96]
a massively parallel genetic algorithm for image analysis (MPGAIA)
which run in a DAP 510 machine. This algorithm is similar to that of Manderick
and Spiessens [MS89].
The main difference is the inclusion of real valued
genes instead of grey coded binary genes of the model of Manderick
and Spiessens.
- Shapiro and Naveta [SN94]
used a fine-grained GA algorithm to predict the secondary structure of RNA.
They used the native X-Net topology of the MasPar-2
computer to define the neighborhood of each individual. The X-Net topology
connects a processing element to eight other processors. Different logical
topologies were tested
with the GA: all eight nearest neighbors, four nearest
neighbors, and all neighbors within a specified distance d.
The results for d>2 were the poorest, and the
four-neighbor scheme consistently outperformed the topology with eight neighbors.
- Kohlmorgen et al. studied in [KSH96]
the island and some variants of the neighborhood model on the massively parallel
computer MasPar MP1 with 16K processors.